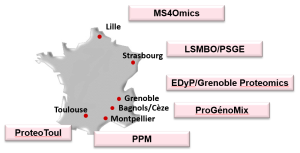
EDyP
Exploring the Dynamics of ProteomesGrenoble
LSMBO
BioOrganic Mass Spectrometry LaboratoryStrasbourg
ProTeoToul
Proteomics and Mass Spectrometry of BiomoleculesToulouse
ProGénoMix
ProGénoMix platformBagnols-sur-Cèze
PPM
Pôle ProtéomeMontpellier
MS4Omics
Mass Spectrometry Platform for OmicsLille
ProFI facilities are distributed over six sites in France
Why Choose our theme?
Set your website in 5 minutes with ready Designs!
The Proteomics French Infrastructure is a national project set up to provide a unified service platform in proteomics studies. It's open to both public and private collaborations.
Proteomics Services
make your work easier and faster!

Large scale protein identification and quantification
Current proteomics methods and instrumentation represent today a powerful tool to get new insights into complex biological systems.

Targeted quantification
Targeted strategies have emerged ten years ago and are best suited when precise, idealy absolute, quantification of finite sets of proteins are required with highest sensitivity.

Post-translational modifications: detection and quantification
Proteins are very often the subject of various post-tanslational modifications (PTMs) that can impact their association with other proteins and globally other biomolecules, their subcellular localization ans their stability.

Interactomics and protein complexes
Interactomics and protein complexes investigations are also based on our strategies and instrumentation for large scale and targeted quantification.

Characterization of intact proteins and complexes
The next frontiers in proteome research are the study of protein-ligand interactions, protein-peptide and protein-protein complexes, together with entire protein interaction networks by high-throughput methods.

Large scale protein identification and quantification
Current proteomics methods and instrumentation represent today a powerful tool to get new insights into complex biological systems.

Targeted quantification
Targeted strategies have emerged ten years ago and are best suited when precise, idealy absolute, quantification of finite sets of proteins are required with highest sensitivity.

Post-translational modifications: detection and quantification
Proteins are very often the subject of various post-tanslational modifications (PTMs) that can impact their association with other proteins and globally other biomolecules, their subcellular localization ans their stability.

Interactomics and protein complexes
Interactomics and protein complexes investigations are also based on our strategies and instrumentation for large scale and targeted quantification.

Characterization of intact proteins and complexes
The next frontiers in proteome research are the study of protein-ligand interactions, protein-peptide and protein-protein complexes, together with entire protein interaction networks by high-throughput methods.
Key Figures
Last News
 Training
Training Workshop on Quantitative Proteomics 2024
ProFI organizes a new edition of the CNRS workshop (Ecole Thématique, held in French) in quantitative proteomics with a focus on data...
 Events
Events MetaboHUB and ProFI Joint Day
The MetaboHUB and ProFI national infrastructures are organizing a joint day in Toulouse on Tuesday, March 26, 2024. During the morning, 3...
 Events
Events Welcome to ProFI’s new nodes !
Following the final decision of our Institutional Committee on September 20th 2023, we are delighted to announce that three new nodes are...








